Research
We use computational tools to study the relationship between genotype and cancer-associated phenotypes at multiple scales.
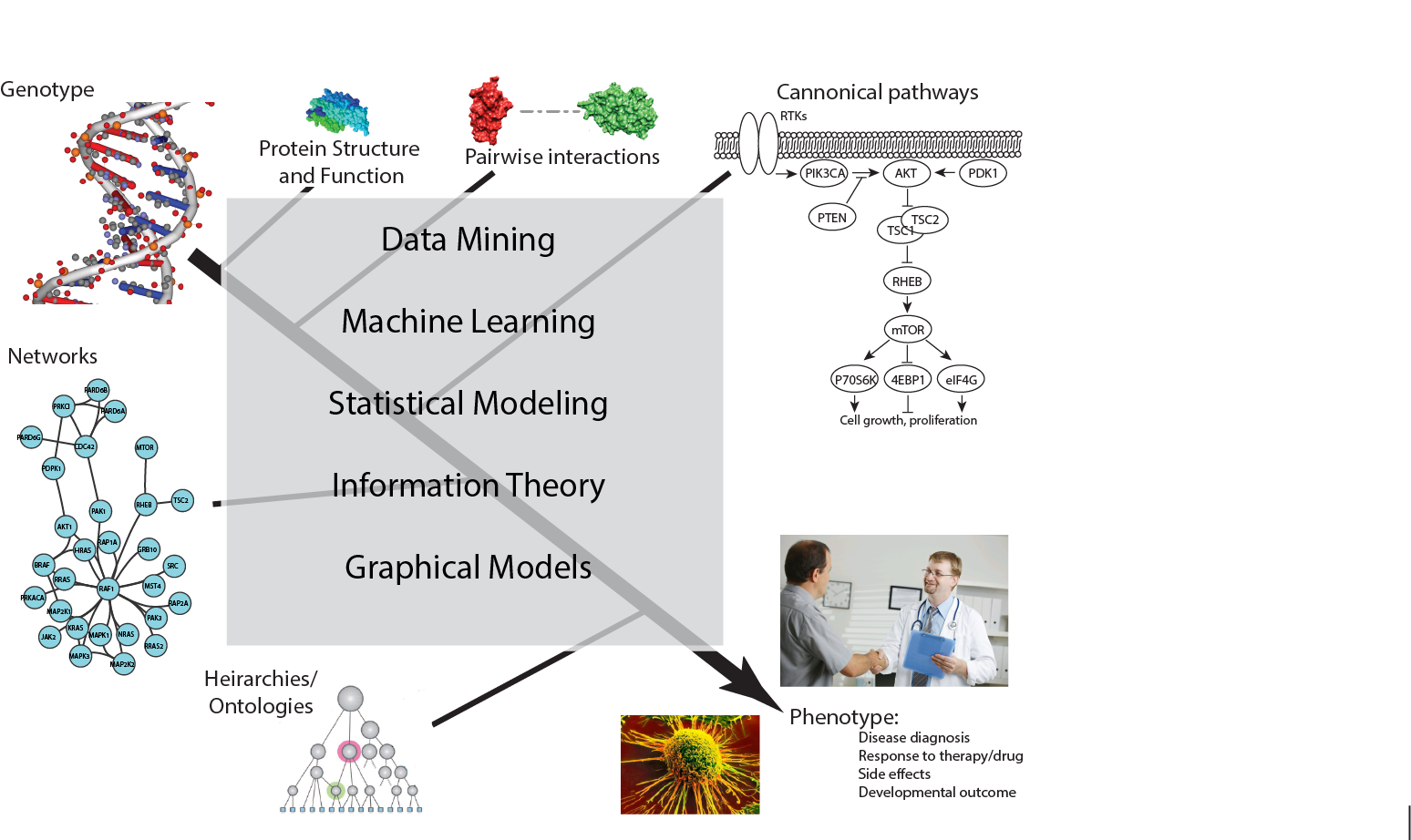
Studying the impact of cancer mutations on intracellular biological activities
Tumors develop through the accumulation of genetic and epigenetic alterations that confer an inappropriate growth advantage to tumor cells. Cancer-causing mutations target specific intracellular biological activities related to cell survival, growth, differentiation and genome maintenance. However no two tumors share the identical set of mutations, suggesting that there are many different ways to impact the same cellular behaviors. We are developing network models to investigate how different mutations converge on neoplastic cellular behaviors. The knowledge gained from these models will help inform future efforts to develop effective cancer therapies.

Related publications:
Engin B, Carlin D, Pratt D, Carter H Modeling of RAS Complexes Supports Roles in Cancer for Less Studied Partners BMC Biophysics Aug 11;10(1):5 (2017)
Engin B, Kreisberg J, Carter H* Structure-based Analysis Reveals Cancer Missense Mutations Target Protein Interaction Interfaces PLoS One Apr 4;11(4):e0152929 (2016)
Engin B, Hofree M, Carter H* Identifying Mutation-Specific Cancer Pathways Using a Structurally Resolved Protein Interaction Network Pac Symp Biocomput. 84-95 (2015)
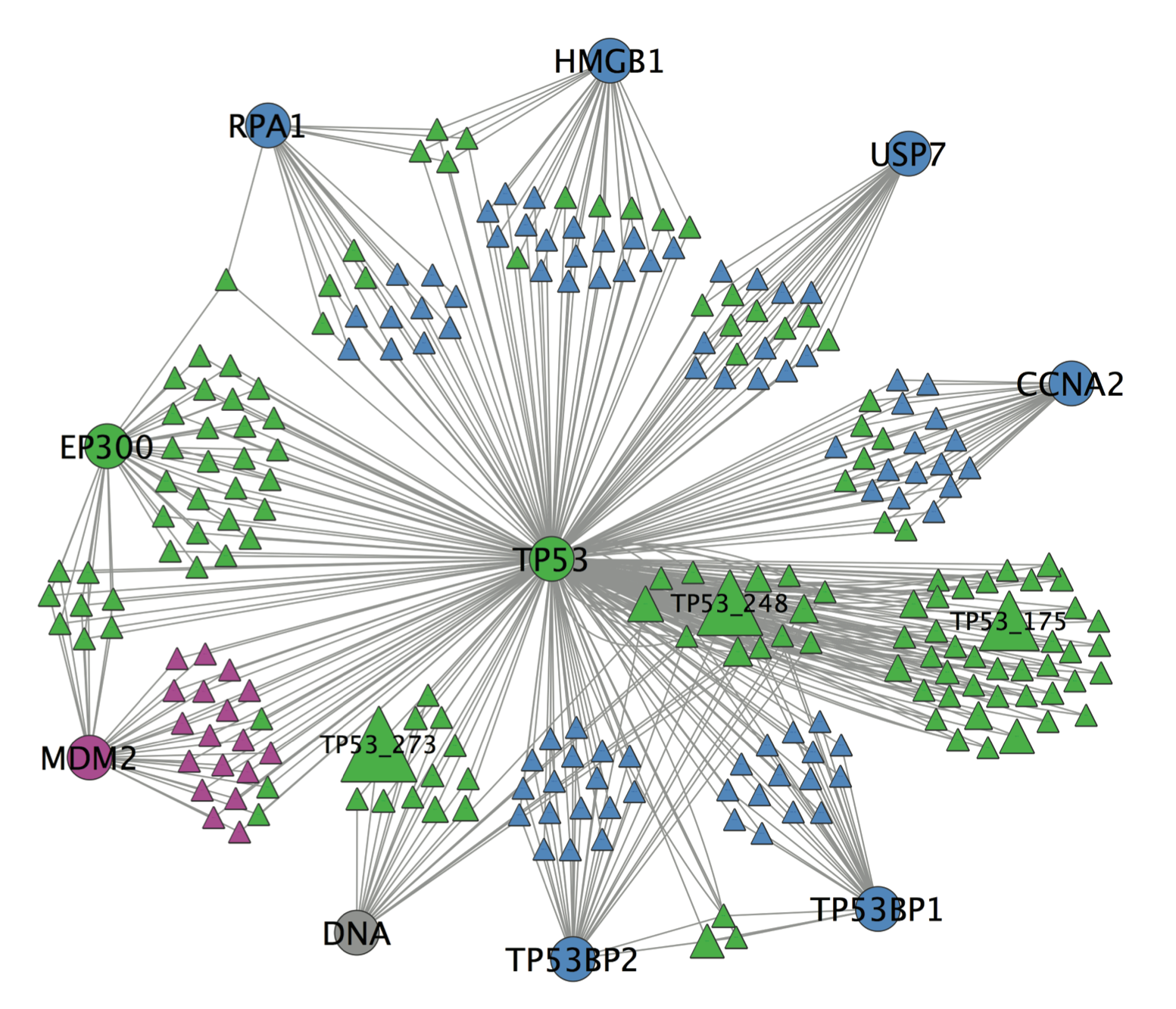
Discriminating cancer drivers from passengers
Each cancer genome is characterized by tens-to-thousands of DNA mutations, but only a minority of those mutations is expected to drive tumor development and progression. The vast majority of mutations are so called “passenger” mutations that are carried along in the genomes of proliferating tumor cells. Discriminating drivers from passengers remains an important challenge in cancer ‘omics research. We are developing new strategies to improve identification of drivers by studying mutations in the context of protein interaction networks.
Related Publications:
Carter H, Samayoa J, Hruban RH, Karchin R, Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM), Cancer Biol Ther 10, 582, Sep (2010)
Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R, Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations, Cancer Res 69, 6660, Aug 15 (2009)
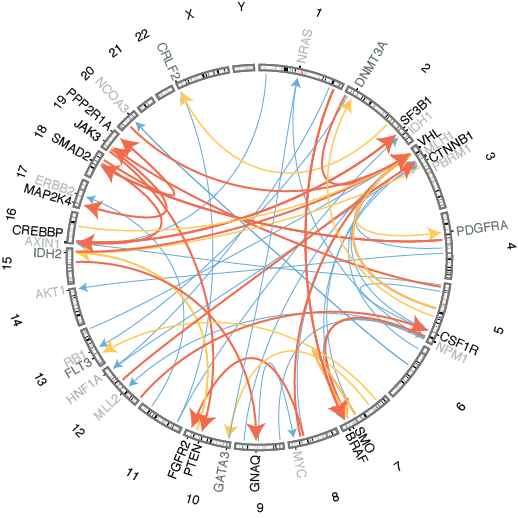
Uncovering interactions between the inherited genome and tumor development
Cancer develops on an individual’s unique genetic background. While some inherited mutations are known to predispose individuals to developing tumors, little is known about the effects of such inherited genetic variation on the evolution of the tumor genome. We are working to uncover new regions of the genome that influence the probability that certain mutations will occur during tumor development. We previously described multiple loci that influence mutation rates of known cancer genes and the site at which a tumor will occur.
Related publications:
Marty R, Kaabinejadian S, Rossell D, Slifker MJ, van de Haar J, Engin HB, de Prisco N, Ideker T, Hildebrand WH, Font-Burgada J, Carter H MHC-I genotype restricts the oncogenic mutational landscape Cell (2017)
Carter H, Marty R, Hofree M, Gross A, Jensen J, Fisch K, Wu X, DeBoever C, Van Nostrand E, Song Y, Wheeler E, Kreisberg J, Lippman S, Yeo G, Gutkind S, Ideker T Widespread interacion of inherited polymorphisms with somatic events in cancer Cancer Discovery Apr 1;7(4):410-23 (2017)